Data description and preprocessing
Our study compiled a comprehensive dataset from Shenzhen People’s Hospital, consisting of 4,352 vitiligo patients and 28,270 healthy controls, collected between 2019 and 2022. The experimental data in this study were collected following diagnosis initial stage of vitiligo. Informed consent was obtained from all participants and the study adhered to strict confidentiality and ethical standards. The research methods were approved by the Institutional Review Board of Shenzhen People’s Hospital, Second Clinical Medical College of Jinan University, China (IRB #LL-KY-2023120-01), ensuring compliance directives and regulations in force.
Kumar et al.6 highlighted the challenges of collecting mixed vitiligo due to its rarity. In our study, emphasis was placed on the differences between segmental and non-segmental vitiligo, excluding consideration of mixed vitiligo. To improve data quality, we performed various preprocessing steps including data selection, column renaming, removing duplicates, handling missing values, ensuring consistency, sorting and correcting values aberrant. Although dimensionality reduction techniques such as Lasso Regression and Elastic Network simplify complex data, they caused imbalances in our study data, prompting us to exclude them. Instead, we used random oversampling to effectively address multiclass imbalance, thereby improving the reliability of our analysis in this vitiligo patient dataset.
Proposal Overview
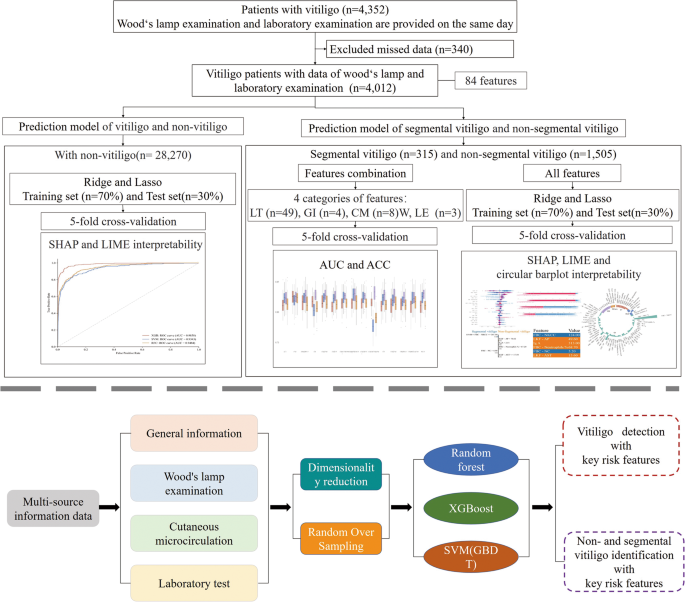
This dataset included 84 clinical features, detailing demographic, gender, age and subtype diagnoses (Supplementary Table S1 and Supplementary Table S2). These features were categorized into four groups: general information (GI), laboratory tests (LT), Wood’s lamp examination (WLE), and comprehensive cutaneous microcirculation (CM) terminology, as shown in Supplementary Table S3 . Leveraging this rich clinical data, we designed a three-step approach for our analysis: first, predicting vitiligo diagnosis and assessing risks; second, classify vitiligo subtypes; and third, to evaluate the importance of different feature categories for the classification of segmental and non-segmental vitiligo (as shown in Fig. 1). We used machine learning techniques to predict vitiligo compared to healthy populations, developed predictive models for segmental and non-segmental vitiligo, and elucidated critical features contributing to our risk assessments. These diagnostic criteria, used by professional dermatologists, are an important tool for confirming cases of vitiligo. Furthermore, we systematically categorized all features and evaluated their individual contributions to the classification of segmental and non-segmental vitiligo.
A graphic illustration of this study.
Feature selection using machine learning
In our study, we examined several machine learning algorithms, including Random Forest (RF), XGBoost, Support Vector Machine (SVM), and Gradient Boosting Decision Trees (GBDT), each offering distinct advantages for different applications.22. We split our dataset in a ratio of 7:3 for the training and testing sets, maintaining the independence of the testing set to ensure unbiased evaluation. Using random 5-fold cross-validation in the training set, we evaluated the accuracy and area under the curve (AUC) values for a comprehensive comparison of the algorithms.
Additionally, we identified a significant data imbalance in the classification of vitiligo patients versus healthy individuals and between segmental and non-segmental vitiligo groups, with positive/negative data ratios of 85:15 and 17:83, respectively (Supplementary Table S1 and Supplementary Table S2). To mitigate this imbalance, we used random walk oversampling, as shown in reference23. This technique involves replicating instances of the underrepresented class to achieve a more balanced class distribution, thereby improving the model’s ability to generalize across various data representations.
RF is deeply rooted in decision trees, where each tree constitutes a fundamental part of its predictive power. However, what really sets RF apart is its ingenious integration of feature randomness throughout the tree building process. This comprehensive approach encompasses key steps including random sampling, feature selection, tree construction, and the final step of averaging predictions.24. Meanwhile, XGBoost takes a different path, using a meticulous and greedy algorithm to deeply explore potential split points within the dataset’s features. At each split, XGBoost calculates the gains and, through an iterative process, selectively identifies the most promising splits while maintaining tight control over tree depth to avoid overfitting.25.
SVM, on the other hand, starts as a linear classifier but has a remarkable ability to transcend linearity thanks to kernel trickery. The quest of SVM is to discover a hyperplane in the feature space that not only accurately classifies the training data, but also maximizes the margin, ultimately defining the problem as a convex optimization challenge.26. Meanwhile, GBDT takes a staged approach to minimize errors. In each iteration, GBDT adjusts the gradients and approximates the residuals following the direction of the negative gradient of the loss function. GBDT relies on Classification And Regression Tree (CART) regression trees at each step and, crucially, accumulates the results from all these trees. This strategy makes GBDT remarkably adaptable to classification tasks, even if the constituent trees remain regression trees rather than classification trees.27.28.
To robustly evaluate the performance of these various machine learning methods, our study uses the widely recognized technique of random 5-fold cross-validation. In this approach, the training dataset is conveniently divided into 5 subsets, with 4 subsets randomly selected for training purposes while the remaining subset is dedicated for testing. This process is repeated 5 times and the average test error serves as a holistic measure of generalization error.29. Importantly, this methodology ensures that each training sample serves as both a training and testing data point, effectively improving the usability of the datasets and providing rigorous evaluations of the selected algorithms.
Performance measures
To better compare the effect and performance of different methods, we consider three performance metrics: confusion matrix, precision, and AUC value for experimental comparison.
$$\:Acc=\:\frac{\text{T}\text{P}+\text{T}\text{N}}{\text{T}\text{P}+\text{T} \text{N}+\text{F}\text{P}+\text{F}\text{N}}$$
where TP denotes the true positive case, FP denotes the false positive case, FN denotes the false negative case and TN denotes the true negative case.
$$\:AUC=\:\frac{\sum\:I({P}_{Positive},\:{p}_{negative})}{M\text{*}N}$$
Interpretability of the model
In recent research, explainable artificial intelligence (XAI) has emerged as a crucial tool in the biomedical field, providing transparent and interpretable models that help understand complex data-driven decisions.30.31. In the complex context of a multi-source dataset, we hope to provide a more nuanced explanation. Explanations of SHapley Additives (SHAP)32 and interpretable local explanations independent of the model33 allow us to understand the impact of each feature on individual predictions of model results, i.e. providing clear and actionable insights into the distinction between healthy individuals and vitiligo patients, as well as as segmental and non-segmental vitiligo, where understanding each specific impact of features is crucial.
SHAP is a technique designed to elucidate how a model, such as XGBoost, arrives at its predictions by quantifying the influence of each attribute within an observation “x”. It does this by dissecting individual predictions into their respective feature contributions. The mathematical representation of these SHAP values is as follows:
$$\:f\left(x\right)=\:g\left(z^\prime\right)=\:{\varnothing\:}_0+\:\sum\limits_{i=1}^M{ \varnothing\:}_{i}z^{\prime}_{i}$$
where f(x) denotes the predicted value of the sample in the decision tree;\(\:\:{\mathbf{z}}_{\mathbf{i}}^{\mathbf{{\prime\:}}}\:\in\:{\{0,\:1\} }^{\mathbf{M}}\) indicates the number of features included in the decision path where this sample is found among all features; And \(\:{{\varnothing}}_{\text{0}}\) indicates that the SHAP value of the feature is 0 if the feature is not in the decision path. The SHAP value is calculated as follows:
$$\:{{\varnothing}}_{\text{i}}=\:{\sum\:}_{\text{S}\subseteq\:\text{N}\left\{\text{ i}\right\}}\frac{\left|\text{S}\right|!\left(\text{M}-\left|\text {S}\right|-1\right) !}{\text{M}!}\left({\text{f}}_{\text{x}}\right(\text{S}\cup\ :\left\{\text{i}\ right\}-{\text{f}}_{\text{x}}\left(\text{S}\right))$$
where N denotes the set of all features in the training set; M designates the dimension; S designates the subset extracted from N; \(\:\left|\varvec{S}\right|\) denotes the dimensions extracted from the subset. Compared to other feature assignment methods, SHAP values also satisfy a desirable set of properties. To derive the SHAP values, we trained the dataset of all observed diagnosed vitiligo cases and generated SHAP plots of the top 14 feature values that influenced the vitiligo prediction of the to better understand the performance of the ‘.
To better understand the behavior of a specific target feature, such as a patient, LIME (Local Interpretable Model-agnostic Explanations) uses a locally trained surrogate model. It is important to note that LIME does not rely on a specific interpretation method to approximate the target black box predictions. This approach leads to the following representation:
$$\:{\upxi\:}\left(\text{x}\right)=\text{a}\text{r}\text{g}\text{m}\text{i}\text{ n}\:\text{L }\left(\text{f},\:\text{g},\:{{\uppi\:}}_{\text{x}}\right)+{\ Omega\:}\left(\text{g}\right)$$
where f designates the original, that is to say what must be explained; g designates simple; G is a set of, such that all possibilities; \(\:{{\uppi\:}}_{\text{x}}\) denotes the difference between the data x’ in the new dataset and the original data instance x; and Ω(g) denotes the complex degree. In the study, we explained the target characteristics of each patient. The prediction probability graph shows the predicted value for the sample, the importance weight of each variable in the sample, and the value of each variable. Additionally, we can see the prediction that the patient has vitiligo through this sample.